 –
– |
All times below are listed in Eastern Time (US)
|
Sensors for biological measurements could change the way that we currently measure analytes in the body, but the design requirements for implementing these tools in vivo are challenging. Among the obstacles that sensors face include fouling, specificity, and biocompatibility/foreign body response. A variety of sensors, including electrochemical and optical, have progressed significantly to address these challenges. This symposium will highlight sensing technologies that have made major advances in long-term sensing in vivo.
|
 09:00 – 11:35
09:00 – 11:35 |
Day 1: Nanomaterials in Sensing ACS Sensors
|
Sensors for biological measurements could change the way that we currently measure analytes in the body, but the design requirements for implementing these tools in vivo are challenging. Among the obstacles that sensors face include fouling, specificity, and biocompatibility/foreign body response. A variety of sensors, including electrochemical and optical, have progressed significantly to address these challenges. This symposium will highlight sensing technologies that have made major advances in long-term sensing in vivo.
|
 09:00 – 09:10
09:00 – 09:10 |
Opening Remarks |
Sensing Science in Brain Chemistry
Development of new sensing strategies and methodologies to directly, selectively, and sensitively record chemical signals of neurons during brain functions has drawn increasing attention because information on the dynamics of chemical signals is very essential to understanding the chemical essence involved in brain functions, for example, neurotransmission and diagnosis and therapy of brain diseases. However, the chemical and physiological complexity of the central nervous system (CNS) unfortunately make this pursuit very challenging to the conventional sensing/analytical protocols. Aiming at this challenge, we have been working on sensing science in live brain ranging from mechanistic development (mainly with rationally modulating electrode/brain interface) to in vivo understanding brain chemistry. This topic will focus on our recent attempts on sensing science in live brain based on rational design and regulation of electrode/brain interface and its application for in vivo understanding brain chemistry.
|
 09:10 – 09:40
09:10 – 09:40 |
Keynote Lecture: Lifelong Sensors for Overall Health Monitoring and Prevention Hossam Haick, Israel Institute of Technology
|
Lifelong Sensors for Overall Health Monitoring and Prevention
Contemporary medicine suffers from many shortcomings in terms of successful disease diagnosis and treatment, both of which rely on detection capacity and timing. The lack of effective, reliable, and affordable detection and real-time monitoring limits the affordability of timely diagnosis and treatment. A new frontier that overcomes these challenges relies on smart health monitoring systems that combine wearable sensors and an analytical modulus. The current talk presents recent advances from our team on smart biocompatible and self-healing materials for the development of multifunctional sensors and wearable devices for an overall health monitoring and prevention while providing a bird’s eye-view of their characteristics, functions, and applications. The talk also shows how these wearables could be fitted with artificial intelligence (AI) to support systems for clinical decision in early detection, accurate diagnosis, and long-life monitoring of health disorders. The ongoing challenges and future prospects for providing personal healthcare with AI-assisted support systems relating to clinical decisions will be also presented and discussed.

Hossam Haick
Department of Chemical Engineering and Russell Berrie Nanotechnology Institute, Technion – Israel Institute of Technology
Email: hhossam@technion.ac.il
lnbd.technion.ac.il
|
 09:40 – 10:10
09:40 – 10:10 |
Keynote Lecture: Real-time Biosensor: Continuous Measurements of Biomolecules in Live Subjects H. Tom Soh, Stanford University
|
Real-time Biosensors: Continuous measurements of biomolecules in live subjects
A biosensor capable of continuously measuring specific molecules in vivo would provide a valuable window into patients’ health status and their response to therapeutics. Unfortunately, continuous, real-time molecular measurement is currently limited to a handful of analytes (i.e. glucose and oxygen) and these sensors cannot be generalized to measure other analytes. In this talk, we will present a biosensor technology that can be generalized to measure a wide range of biomolecules in living subjects. To achieve this, we develop synthetic antibodies (aptamers) that change its structure upon binding to its target analyte and produce an electrochemical current or emit light. Our real-time biosensor requires no exogenous reagents and can be readily reconfigured to measure different target analytes by exchanging the aptamer switches in a modular manner. Importantly, we will discuss methods for generating the aptamer switches which are at the heart of this biosensor technology.

H. Tom Soh
Professor of Electrical Engineering and Radiology, Stanford University
Email: tsoh@stanford.edu
https://sohlab.stanford.edu/
|
 10:10 – 10:40
10:10 – 10:40 |
Keynote Lecture: Nanomaterials for Implantable and Wearable Continuous Monitoring Anne Andrews, University of California, Los Angeles
|
Nanomaterials for Implantable and Wearable Continuous Monitoring
New technologies precede and enable discoveries in biology and medicine. We seek to discover how neurotransmitters encode emotion-related information. However, poor chemical, spatial, and temporal resolution have limited dynamic measurements of brain chemistries limiting dynamic data acquisition. To understand chemical signaling in the brain and other physiological compartments at scales pertinent to encoded information, micro- to nanoscale sensors are needed for multiplexed and highly selective readouts with fast response times. We design, develop and deploy sensors that approach these critical attributes. We have developed electronic biosensors to investigate chemical signaling. Target recognition is by oligonucleotide receptors (aptamers) linked to nanometer thin-film field-effect transistor (FET) arrays for transducing binding events. We have selectively detected small molecule targets including serotonin, dopamine, glucose, phenylalanine, and cortisol in a label-free manner in undiluted physiological fluids over 56 orders of magnitude with nMfM detection limits. We have deployed neuroprobes based on stiff and flexible substrates and monitored serotonin in the brain and spinal cord at physiological concentrations. We have also monitored stress- and circadian-related changes in cortisol in sweat via a fully autonomous smartwatch. Currently, we are focused on multiplexing to enable integrated physiological monitoring.

Anne Andrews
Professor of Psychiatry & Biobehavioral Sciences and Chemistry & Biochemistry; University of California, Los Angeles
Email: AAndrews@mednet.ucla.edu
https://serotonin.ucla.edu/
|
 10:40 – 10:45
10:40 – 10:45 |
Interactive Break |
Engineering the Nanoparticle Corona and Optical Techniques for In-Vivo Sensing at Biological Interfaces
Nanosensors, particularly those based on nanoparticle transducers, have demonstrated the ability to detect the binding of molecules at the single analyte level. Our lab at MIT has been interested in how the nanoparticle corona – the region of adsorbed molecules surrounding the particle surface – can be engineered for molecular recognition. We have recently introduced a method we call CoPhMoRe or Corona Phase Molecular Recognition1 for discovering synthetic, heteropolymer corona phases that form molecular recognition sites at the nanoparticle interface, selected from a heteropolymer library. We show that certain synthetic heteropolymers, once constrained onto a single-walled carbon nanotube by chemical adsorption, also form a new corona phase that exhibits highly selective recognition for specific molecules. We have a growing list of biomolecules that we can detect using this approach including riboflavin, L-thyroxine, dopamine, nitric oxide, sugar alcohols, estradiol, as well as proteins such as fibrinogen. The results have significant potential in light of the fact that nanoparticles such as single walled carbon nanotubes can be interfaced to biological systems at the sub-cellular level, with unprecedented sensitivity. Several recent demonstrates indicate that spatial and temporal information on cellular chemical signaling can be obtained using arrays of such sensors. To enable the use of these types of sensors, we have developed techniques that allow optical excitation to propagate through the in-vivo environment, avoiding unfavorable tissue scattering and intrinsic autofluorescence. We develop a wavelength-induced frequency filtering (WIFF) whereby the fluorescence excitation wavelength is modulated across the absorption cross-section of the fluorescent bisensor, allowing the emission signal to be separated from the autofluorescent background, increasing the desired signal relative to noise, and internally referencing it to protect against artifacts. Using highly scattering tissue phantoms, an SKH1-E mouse model, and other complex tissue types, we show that WIFF significantly improves the in vivo signal to noise ratio (SNR) of fluorescent sensors up to 52 fold for commonly employed chromophores in the visible spectrum. WIFF extends measurements to extremely deep implants up to 5.5±0.1 cm depth in chicken breast tissue when using probes excited at 730 nm and emitting between 1150 and 1300 nm, even allowing the monitoring of riboflavin diffusion in thick tissue. As an application, we demonstrate that WIFF enables the detection of the chemotherapeutic activity of temozolomide trans-cranially 2.4±0.1 cm through the porcine brain without the use of fiber optic or cranial window insertion. These results based on WIFF open up new avenues of biomedical research by extending a large number of fluorescent sensors to previously inaccessible in vivo environments.
|
 10:45 – 11:30
10:45 – 11:30 |
Plenary Lecture Ali Javey, UC Berkeley
|
Engineering the Nanoparticle Corona and Optical Techniques for In-Vivo Sensing at Biological Interfaces
Nanosensors, particularly those based on nanoparticle transducers, have demonstrated the ability to detect the binding of molecules at the single analyte level. Our lab at MIT has been interested in how the nanoparticle corona – the region of adsorbed molecules surrounding the particle surface – can be engineered for molecular recognition. We have recently introduced a method we call CoPhMoRe or Corona Phase Molecular Recognition1 for discovering synthetic, heteropolymer corona phases that form molecular recognition sites at the nanoparticle interface, selected from a heteropolymer library. We show that certain synthetic heteropolymers, once constrained onto a single-walled carbon nanotube by chemical adsorption, also form a new corona phase that exhibits highly selective recognition for specific molecules. We have a growing list of biomolecules that we can detect using this approach including riboflavin, L-thyroxine, dopamine, nitric oxide, sugar alcohols, estradiol, as well as proteins such as fibrinogen. The results have significant potential in light of the fact that nanoparticles such as single walled carbon nanotubes can be interfaced to biological systems at the sub-cellular level, with unprecedented sensitivity. Several recent demonstrates indicate that spatial and temporal information on cellular chemical signaling can be obtained using arrays of such sensors. To enable the use of these types of sensors, we have developed techniques that allow optical excitation to propagate through the in-vivo environment, avoiding unfavorable tissue scattering and intrinsic autofluorescence. We develop a wavelength-induced frequency filtering (WIFF) whereby the fluorescence excitation wavelength is modulated across the absorption cross-section of the fluorescent bisensor, allowing the emission signal to be separated from the autofluorescent background, increasing the desired signal relative to noise, and internally referencing it to protect against artifacts. Using highly scattering tissue phantoms, an SKH1-E mouse model, and other complex tissue types, we show that WIFF significantly improves the in vivo signal to noise ratio (SNR) of fluorescent sensors up to 52 fold for commonly employed chromophores in the visible spectrum. WIFF extends measurements to extremely deep implants up to 5.5±0.1 cm depth in chicken breast tissue when using probes excited at 730 nm and emitting between 1150 and 1300 nm, even allowing the monitoring of riboflavin diffusion in thick tissue. As an application, we demonstrate that WIFF enables the detection of the chemotherapeutic activity of temozolomide trans-cranially 2.4±0.1 cm through the porcine brain without the use of fiber optic or cranial window insertion. These results based on WIFF open up new avenues of biomedical research by extending a large number of fluorescent sensors to previously inaccessible in vivo environments.
|
 11:30 – 11:35
11:30 – 11:35 |
Closing Remarks |
|
 –
– |
All times below are listed in Eastern Time (US)
|
The SARS-CoV-2 pandemic demonstrates the need for measurement tools for rapid and accurate diagnostics, remote measurements and fundamental studies on virus structure and function. This symposium highlights recent advances in sensing, microfluidics, sequencing, mass spectrometry and optical spectroscopy applied to virus diagnostics and characterization with an emphasis on SARS-CoV-2.
|
 09:00 – 11:35
09:00 – 11:35 |
Day 2: Across Chemistry, Space and Time: Single Cell Characterization from the Chemical Perspective Analytical Chemistry
|
The SARS-CoV-2 pandemic demonstrates the need for measurement tools for rapid and accurate diagnostics, remote measurements and fundamental studies on virus structure and function. This symposium highlights recent advances in sensing, microfluidics, sequencing, mass spectrometry and optical spectroscopy applied to virus diagnostics and characterization with an emphasis on SARS-CoV-2.
|
 09:00 – 09:10
09:00 – 09:10 |
Opening Remarks |
Solid-State Nanopore Platform Integrated with Machine-Learning for Digital Diagnosis of Virus Infection
The variability of bioparticles remains a key barrier to realizing the competent potential of nanoscale detection into a digital diagnosis of an extraneous object that causes an infectious disease. We developed label-free virus identification based on machine-learning classification. Single virus particles were detected using nanopores, and resistive-pulse waveforms were analyzed multilaterally using artificial intelligence. In the discrimination, over 99% accuracy for five different virus species, including influenza and corona virus species, was demonstrated. This advance is accessed through the classification of virus-derived ionic current signal patterns reflecting their intrinsic physical properties in a high-dimensional feature space. Moreover, consideration of viral similarity based on the accuracies indicates the contributing factors in the recognitions. We also developed a portable robust ionic current sensor using a bridge circuit that offers a high signal-to-noise (S/N) ratio by suppressing background current as a useful tool for detecting sub- to several-micron scale particles such as virus and bacteria. Because the portable robust ionic current sensor can tolerate increased noise in current sensing, a simple, lightweight electromagnetic shield can be used and measurements under large electromagnetic noise conditions can be made. This sensor combined with machine-learning system enables us to identify several virus species and several bacteria with over 90 % accuracy.
|
 09:10 – 09:40
09:10 – 09:40 |
Keynote Lecture: Electrochemical Cytometry of Nanovesicles at Nanotip and Nanopore Electrodes Andy Ewing, University of Gothenburg
|
Electrochemical Cytometry of Nanovesicles at Nanotip and Nanopore Electrodes
In recent years, nano-electrochemical approaches have been developed to realise the quantitative measurement of intravesicular content and real-time monitoring of their release dynamics. My group has developed new techniques based first on capillary electrophoresis followed by vesicle impact electrochemical cytometry (VIEC), and subsequently stochastic VIEC and intracellular VIEC (IVIEC) to provide a highly effective way to quantify the electroactive contents inside cellular vesicles. This has allowed direct comparison of the quantity of molecules released by exocytosis to those in the vesicles. Partial release is observed across all cell types examined to date by electrochemical methods and appears to be regulated. We have used these methods and imaging mass spectrometry to directly examine plasticity that might lead to the formation of initial memory and a new approach with both intracellular and extracellular measurements at the same time has led to time resolved measurements of plasticity in real time. This plasticity is apparent as a change in fraction released which is observed in drug-treated cells.
We have also recently developed two new methods using resistive nanopores and nanopipette electrodes to simultaneously measure or sort vesicle size and transmitter content, allowing us to correlate individual vesicle size with content and then calculate concentration. In one approach, we have combined resistive pulse sensing with VIEC to simultaneously measure size and content of isolated nanometer chromaffin vesicles. In the other approach we have used specifically sized nanopipette electrodes to narrowly sort vesicle size and measure content in both VIEC and IVIEC.
We have also used our cytometry method to examine stress granules (SGs), membraneless subcellular organelles that assemble to capture mRNAs and proteins during stresses including oxidative stress, heat shock, viral infection, proteasomal inhibition, ER (endoplasmic reticulum) stress, UV irradiation, among others. We used nanotip electrochemical cytometry to determine that reactive oxygen species (ROS) are generated inside SGs. Although the exact biological function of SGs remains to be discovered, our work unveils a correlation between SG biology and pathogenesis in neurodegeneration and some cancers.
In a new chemical approach, we have discovered that the chaotropic anion (SCN-) creates a two-step process (around 30% doublet peaks) when examining adrenal chromaffin vesicles with VIEC. We have then used this to independently count molecules in each subvesicular compartment, the halo and protein dense-core, and shown the fraction of catecholamine binding to the dense-core is 68%. Moreover, we differentiated two distinct populations of large dense-core vesicles and quantified their content, which might correspond to immature (43%) and mature (30%) large dense-core vesicles, to reveal differences in their biogenesis.

Andrew Ewing
Professor of Chemistry and Molecular Biology, Univ. of Gothenburg
Email: Andrewe@chem.gu.se
http://andrewewinggroup.se/members/andrew-ewing/
|
 09:40 – 10:10
09:40 – 10:10 |
Keynote Lecture: Watching a Molecular Orchestra in a Living System via Bond-Selective Microscopy Ji-Xin Cheng, Boston University
|
Watching a molecular orchestra in a living system via bond-selective microscopy
Most of our knowledge about chemistry inside a cell is from in vitro analysis of biomolecules in solutions. Probing the spatial and temporal dynamics of molecular interactions is essential to understanding the machinery of life. Chemical microscopy based on spectroscopic signals from chemical bond vibrations opens a new window to watch the life at molecular level with sub-micron spatial resolution. Two advanced chemical microscopies, based on coherent Raman scattering [1] and mid-infrared photothermal effect [2], have allowed real time imaging of structural and reaction chemistry inside a living system. This presentation will introduce these chemical imaging modalities and applications to seeing the unseen in life science. Examples including the findings of altered metabolism in cancer cells and molecular response of organisms to a treatment.
[1] Ji-Xin Cheng*, Sunney X. Xie*, “Vibrational spectroscopic imaging of living systems: an emerging platform for biology and medicine”, Science, review, 2015, 350: 1054.
[2] Yeran Bai, Jiaze Yin, Ji-Xin Cheng*, “Bond-Selective Imaging by Optically Sensing the Mid-Infrared Photothermal Effect”, Science Advances, review, 2021, 7: eabg1559

Ji-Xin Cheng
Inaugural Theodore Moustakas Chair Professor in Photonics and Optoelectronics, Boston University
Email: jxcheng@bu.edu
http://sites.bu.edu/cheng-group
|
 10:10 – 10:40
10:10 – 10:40 |
Keynote Lecture: Single-Cell Metabolome Analysis with Mass Spectrometry Xinrong Zhang, Tsinghua University
|
Single-Cell Metabolome Analysis with Mass Spectrometry
Understanding cellular heterogeneity is important in the study of biological processes but it was limited by existing analytical tools. Recently single-cell MS achieves subcellular resolution and provide new dimensional insights into the hierarchical processes although several analytical complications still exist. Here I describe a micromanipulation system that combines microextraction with ESI-MS for single-cell metabolites1. In addition, an imaging mass spectrometry with a set of computational algorithms was also developed2, which can display multiscale and multicolor tissue tomography together with identification and clustering of single nuclei by their in situ metabolic fingerprints.
[1 ] Chen, AQ; et al., Single cell mass spectrometry with a robotic micromanipulation system for cell metabolite analysis, IEEE transactions on biomedical engineering, 2022, 69(1), 325-333
[2] Yuan, ZY; et al., SEAM is a spatial single nuclear metabolomics method for dissecting tissue microenvironment, Nature methods, 2021, 18(10), 1223-1232

Xinrong Zhang
Executive Editor, Analytical Chemistry
Professor in Analytical Center, Department of Chemistry, Tsinghua University
Email: xrzhang@mail.tsinghua.edu.cn
http://xrzhanggroup.com/xinrong-zhang
|
 10:40 – 10:45
10:40 – 10:45 |
Interactive Break |
Ultrasensitive Immunoassays for Understanding SARS-CoV-2 Infections
Using single-molecule arrays, we have developed digital ELISAs with 2-3 logs better analytical sensitivity than conventional immunoassays. We have used these assays to measure viral proteins, host antibodies and host inflammatory responses to SARS-CoV-2 infections. The ultrasensitivity allows us to measure proteins earlier during infection. Furthermore, the ability to measure extremely low concentrations enables us to observe aspects of disease pathogenesis that provide mechanistic insight. In this talk, I will describe some of our findings that elucidate the development of immune responses and disease severity, as well as features of the host response to mRNA vaccines.
|
 10:45 – 11:30
10:45 – 11:30 |
Plenary Lecture Petra S. Dittrich, ETH Zurich
|
Ultrasensitive Immunoassays for Understanding SARS-CoV-2 Infections
Using single-molecule arrays, we have developed digital ELISAs with 2-3 logs better analytical sensitivity than conventional immunoassays. We have used these assays to measure viral proteins, host antibodies and host inflammatory responses to SARS-CoV-2 infections. The ultrasensitivity allows us to measure proteins earlier during infection. Furthermore, the ability to measure extremely low concentrations enables us to observe aspects of disease pathogenesis that provide mechanistic insight. In this talk, I will describe some of our findings that elucidate the development of immune responses and disease severity, as well as features of the host response to mRNA vaccines.
|
 11:30 – 11:35
11:30 – 11:35 |
Closing Remarks |
|
 –
– |
All times below are listed in Eastern Time (US)
|
Advancing proteomic technology is improving the ability to study the proteome to learn new biology and to aid in the diagnosis of disease. This symposium will probe new strategies to identify and quantify posttranslational modifications to determine their biological roles. The speakers will encompass bottom- up strategies as well as top down to identify PTMs as well as proteoforms.
|
 09:00 – 11:35
09:00 – 11:35 |
Day 3: Deep Learning in Proteomics: Deep Insight or Deeper Pitfalls? Journal of Proteome Research
|
Advancing proteomic technology is improving the ability to study the proteome to learn new biology and to aid in the diagnosis of disease. This symposium will probe new strategies to identify and quantify posttranslational modifications to determine their biological roles. The speakers will encompass bottom- up strategies as well as top down to identify PTMs as well as proteoforms.
|
 09:00 – 09:10
09:00 – 09:10 |
Opening Remarks |
Most studies have established macrophages as an indispensable key player in both innate immune responses and adaptive immunity. Thus, they are ideal therapeutic targets for diseases such as insulin resistance, cancer, type 2 diabetes, atherosclerosis, and periodontitis. Macrophages are not homogenous, and they are generally categorized into two broad but distinct subsets as either classically activated (M1, pro-inflammatory) or alternatively activated (M2, anti-inflammatory). Recent studies have demonstrated that macrophages produce extracellular traps (ETs). ETs are an immune response by which a cell undergoes “ETosis” to release net-like material, with strands composed of cellular DNA that is studded with histones and cellular proteins. Peptidylarginine deiminase 4 (PAD4) is critically involved in chromatin decondensation during the release of ETs. The aim of our study was to investigate the role of PAD enzymes and its downstream effects, such as the protein citrullination in polarization of macrophages to M1 and M2. We used human monocytic cell line THP-1 differentiated by PMA to macrophages. The cells were further differentiated to proinflammatory M1 and anti-inflammatory M2 macrophages by 10 ng/ml LPS (S. Minnesota) and 20 ng/ml IL-4, respectively, and treated with 100 nM PAD inhibitor – BB-Cl-amidine for 48 h. We measured mRNA expression of different PAD isoforms and M1 and M2 markers upon treatment with BB-Cl-amidine. To identify differentially changed proteins and protein citrullination sites we applied a data independent mass spectrometry method (DIA) and hyper-citrullinated spectral library approach. We determined that PAD inhibitor decreased expression of proinflammatory M1 markers and increased expression of anti-inflammatory M2 markers in THP-1 macrophages. PAD2, the most abundant PAD in macrophages, was downregulated in M2 macrophages. Citrullination seems to play a major role in proinflammatory M1 macrophages with several citrullinated proteins involve in platelet activation, signaling and aggregation. The study showed that PADs and protein citrullination could play a critical role in the differentiation of macrophage and inflammatory response. Furthermore, identified citrullinated proteins, could be the new autoantigen that influence anti-citrullinated protein antibodies and can be used as a new marker for disease detection and progression.
|
 10:10 – 09:40
10:10 – 09:40 |
Keynote Lecture: Machine Learning in Biomarker Discovery: Overlooked Elements of Feature Selection and Dataset Design Heather Desaire, University of Kansas
|
Machine Learning in Biomarker Discovery: Overlooked Elements of Feature Selection and Dataset Design
Looking for disease biomarkers in proteomics datasets is often compared to looking for a needle in a haystack; the number of potential markers that can now be quantified is immense. The challenge for data scientists is to find the needles (true biomarkers), when they are present and to not be misled by potentially promising proteins that turn out to be no more useful for discriminating disease than shiny pieces of hay. The presentation will address this challenge, known as feature selection, from a variety of perspectives. As a cautionary tale, we will show that an improperly executed feature selection strategy leads to identifying disease biomarkers that appear to offer “>90% accuracy”, but are, in fact, no better than flipping a coin for predicting disease. Unfortunately, the improperly executed strategy is a pervasive mistake that currently occurs when researchers combine feature selection methods with proteomics datasets; we will demonstrate how the problem can be avoided. A second vignette will illustrate the unequivocal importance of using racially diverse sample sets in biomarker studies. We will show that biomarker candidates for Alzheimer’s Disease, which may be optimally beneficial for a broad, racially diverse population, can be overlooked if the underlying datasets used to identify them are not racially diverse. Identifying useful disease biomarkers is a formidable challenge, but real successes are more likely when researchers attend carefully to good principles of machine learning and dataset design.

Heather Desaire
Dean’s Professor of Chemistry, University of Kansas
Email: hdesaire@ku.edu
https://desairegroup.ku.edu/people/desaire
|
 09:40 – 10:10
09:40 – 10:10 |
Keynote Lecture: From Protein Identification to Functional Interpretation Isabell Bludau, Max Planck Institute of Biochemistry
|
From protein identification to functional interpretation
A fundamental challenge in the analysis and interpretation of proteomics data is to identify functional relevance. While we are routinely capable to find hundreds of proteins or post-translational modifications (PTMs) that change their abundance across biological conditions, the identification of the few functionally important ones among them mostly remains a manual and laborious task. In my talk I will discuss the challenges and opportunities that machine and deep learning offer to facilitate this process. This includes both the direct application of deep learning models to proteomics data, but also opportunities to integrate additional layers of information. This is exemplified by the recent breakthrough of deep learning for protein structure prediction, that has enabled new avenues of proteomics data integration to systematically prioritize functionally promising PTM sites.

Isabell Bludau
Postdoctoral Fellow in the lab of Prof. Matthias Mann, Max Planck Institute of Biochemistry
Email: bludau@biochem.mpg.de
|
 10:10 – 10:40
10:10 – 10:40 |
Keynote Lecture: Deep learning: From Spectra to Protein to Networks Lars Juhl Jensen, University of Copenhagen
|
Deep learning: from spectra to protein to networks
There can be no doubt that deep learning (DL) has had a huge impact on how we analyze biological data. Most biomedical applications of DL have involved data that can be represented as sequences, vectors, or matrices/images. The use of DL in proteomics has so far been limited and mainly focused on prediction of fragment spectra from peptide sequences. While peptides and expression profiles are sequences and vectors, respectively, there is no obvious representation for raw tandem mass-spectrometry (MS) data, which in my view is the main challenge that has held back the use of DL in proteomics. A second big challenge is the heterogeneity of proteomics data compared to, for example, RNA sequencing data. In my presentation, I will briefly cover some promising DL-based methods, which I believe may help us work around these challenges and bring us from spectra to proteins further to protein networks.

Lars Juhl Jensen
Professor at the Novo Nordisk Foundation Center for Protein Research, University of Copenhagen
Email: lars.juhl.jensen@cpr.ku.dk
https://jensenlab.org/people/larsjuhljensen/
|
 10:40 – 10:45
10:40 – 10:45 |
Interactive Break |
New Technologies for Exploiting the Human Glycoproteome for Cardiovascular Physiology and Personalized Medicine
Cell surface glycoproteins and glycans play critical roles in a range of biological functions and disease processes and may be exploited as biomarkers for precision medicine. Despite their biological relevance and utility, glycoproteins and glycans are often understudied largely due to technical challenges. This presentation will describe our recently developed analytical and bioinformatic solutions that enable rapid identification and quantification of cell surface glycoproteins and glycans from small sample sizes. The application of these new methodologies to address outstanding questions in cardiac biology and disease, with an emphasis on precision medicine, will be described.
|
 10:45 – 11:30
10:45 – 11:30 |
Plenary Lecture: Pitfalls and Insights When Training a Peptide Property Prediction Model Exemplified on Prosit Mathias Wilhelm, Technical University of Munich
|
Pitfalls and insights when training a peptide property prediction model exemplified on Prosit
Deep learning is about to become the state-of-the-art machine learning method to model and predict peptide properties such as fragmentation behavior and retention time. One of such models is Prosit which has been continuously expanded over the last years to supported additional peptide classes and peptide properties. However, machine and particular deep learning is increasingly being connected to the reproducibility crisis in science. In this talk, we share our experience in training Prosit, highlight strategies to avoid common pitfalls and provide insights into training a Prosit model from scratch using custom data.

Mathias Wilhelm
Professor of Computational Mass Spectrometry, Technical University of Munich
Email: mathias.wilhelm@tum.de
http://protrein.eu/supervisor/mathias-wilhelm/
|
 11:30 – 11:35
11:30 – 11:35 |
Closing Remarks - Scientific Session |

Latest Applications of 4D-Proteomics using the timsTOF Pro and recent developments in ultra-high sensitivity and single cell proteomics
This session will discuss the PASEF, dia-PASEF and prm-PASEF methods and explain how these methods allow researchers to perform proteomics faster, with better depth of coverage, better sensitivity, and with ore confidence in their identification and quantitation.
After a summary of these methods, this session will cover some of the latest applications of the timsTOF Pro and 4D-proteomics technology to biomarker discovery and validation, immunopeptidomcs and single cell proteomics.
Topics
– Features and performance of 4D-proteomics on the timsTOF Pro
– PASEF, dia-PASEF, prm-PASEF
– High throughput biomarker discovery and validation
– Ultra-high sensitivity for immunopeptidomics and single cell proteomics
Featured Speaker
Gary Kruppa, Ph.D., Vice President of Proteomics, Bruker Daltonics Inc., has over 30 years of experience in the field of mass spectrometry (MS), having served as a Vice President at Bruker Daltonics for over 20 years.
Kruppa received his Ph.D. in chemical physics from the California Institute of Technology, and his BS from the University of Delaware. Kruppa oversees market and applications development management for Bruker’s innovative solutions for research in proteomics.

|
 14:00 – 16:35
14:00 – 16:35 |
Day 3: The Future of Ion Mobility-Mass Spectrometry in Measurement Science Journal of the American Society for Mass Spectrometry
|

Latest Applications of 4D-Proteomics using the timsTOF Pro and recent developments in ultra-high sensitivity and single cell proteomics
This session will discuss the PASEF, dia-PASEF and prm-PASEF methods and explain how these methods allow researchers to perform proteomics faster, with better depth of coverage, better sensitivity, and with ore confidence in their identification and quantitation.
After a summary of these methods, this session will cover some of the latest applications of the timsTOF Pro and 4D-proteomics technology to biomarker discovery and validation, immunopeptidomcs and single cell proteomics.
Topics
– Features and performance of 4D-proteomics on the timsTOF Pro
– PASEF, dia-PASEF, prm-PASEF
– High throughput biomarker discovery and validation
– Ultra-high sensitivity for immunopeptidomics and single cell proteomics
Featured Speaker
Gary Kruppa, Ph.D., Vice President of Proteomics, Bruker Daltonics Inc., has over 30 years of experience in the field of mass spectrometry (MS), having served as a Vice President at Bruker Daltonics for over 20 years.
Kruppa received his Ph.D. in chemical physics from the California Institute of Technology, and his BS from the University of Delaware. Kruppa oversees market and applications development management for Bruker’s innovative solutions for research in proteomics.

|
 14:00 – 14:10
14:00 – 14:10 |
Opening Remarks |

Latest Applications of 4D-Proteomics using the timsTOF Pro and recent developments in ultra-high sensitivity and single cell proteomics
This session will discuss the PASEF, dia-PASEF and prm-PASEF methods and explain how these methods allow researchers to perform proteomics faster, with better depth of coverage, better sensitivity, and with ore confidence in their identification and quantitation.
After a summary of these methods, this session will cover some of the latest applications of the timsTOF Pro and 4D-proteomics technology to biomarker discovery and validation, immunopeptidomcs and single cell proteomics.
Topics
– Features and performance of 4D-proteomics on the timsTOF Pro
– PASEF, dia-PASEF, prm-PASEF
– High throughput biomarker discovery and validation
– Ultra-high sensitivity for immunopeptidomics and single cell proteomics
BRUKER FM: Far and Mid IR Spectral Range Spectroscopy in One Step
Featured Speaker
Gary Kruppa, Ph.D., Vice President of Proteomics, Bruker Daltonics Inc., has over 30 years of experience in the field of mass spectrometry (MS), having served as a Vice President at Bruker Daltonics for over 20 years.
Kruppa received his Ph.D. in chemical physics from the California Institute of Technology, and his BS from the University of Delaware. Kruppa oversees market and applications development management for Bruker’s innovative solutions for research in proteomics.

|
 14:10 – 14:40
14:10 – 14:40 |
Keynote Lecture: Ion Mobility-Mass Spectrometry for High-Throughout Multi-Omics Kelly Hines, University of Georgia
|
Ion Mobility-Mass Spectrometry for High-Throughout Multi-Omics
There is growing interest and appreciation in the use of mass spectrometry-based multi-omics approaches to study biological processes and diseases from a systems-level perspective. The performance of discovery-level multi-omics typically involves the partitioning of a complex sample in its individual components, followed by thorough analysis of each “ome” under optimized LC and MS conditions. However, these multi-omics experiments must be streamlined before their findings can be implemented into diagnostic or prognostic applications. The rapid gas-phase structural separations afforded by IM-MS provides an opportunity for high-throughput measurements of biological samples containing mixtures of lipids, metabolites, peptides, and other biochemicals. We are developing methods for single injection high-throughput multi-omics based on flow injection analysis (FI) and ion mobility-mass spectrometry (IM-MS). The feasibility and advantages of FI-IM-MS will be demonstrated for the microorganism identification to the species and strain levels using integrated lipidomic and metabolomic features.

Kelly Hines
University of Georgia
Email: Kelly.Hines@uga.edu
www.thehineslab.com
|
 14:40 – 15:10
14:40 – 15:10 |
Keynote Lecture: Unraveling the Steroid-ome with Novel Ion Mobility-Mass Spectrometry Methods Christopher Chouinard, Clemson University
|
Unraveling the Steroid-ome with Novel Ion Mobility-Mass Spectrometry Methods
Steroids make up a structurally diverse biochemical class that can be broken down into subclasses based on their function and point of origin in the human body. For example, sex steroids, produced in the gonads and adrenal glands, include androgens, estrogens, and progestogens and are responsible for biological processes such as sexual differentiation and reproduction. As a class, however, steroids are analytically challenging due to the abundance of structural and stereochemical isomers which can be difficult to differentiate using traditional chromatographic and/or mass spectrometric methods. Over the last decade, IM-MS has emerged as a powerful technique for the analysis of the ‘steroid-ome’ due to the increased confidence in identification afforded by the added mobility separation dimension. Applications ranging from anti-doping to monitoring endocrine diseases have benefited from the additional separation dimension afforded by ion mobility. Herein we will discuss a broad overview of recent advances in IM-MS for steroid analysis, with topics to include targeted quantification of anabolic steroids in athlete urine, computational modeling and machine learning approaches for identifying novel metabolites, and functional group specific reactions for improved resolution.

Christopher Chouinard
Assistant Professor, Chemistry Department, Clemson University
Email: cchouinard@fit.edu
research.fit.edu/chouinard
|
 15:10 – 15:40
15:10 – 15:40 |
Keynote Lecture: Elucidating Biomolecular Structures and Disease Mechanisms with Ion Mobility Mass Spectrometry Thanh Do, University of Tennessee Knoxville
|
Elucidating Biomolecular Structures and Disease Mechanisms with Ion Mobility Mass Spectrometry
The two predominant challenges in measurement science are to gain structural information of transient but biologically active molecules, and to perform many fast, complete, and accurate sampling of volume-limited samples. I will discuss our recent work using ion-mobility mass spectrometry (IM-MS) to capture a sophisticated model of amyloid dodecamer exhibiting selective cellular toxic vulnerability similar to what have been found in Alzheimer’s disease. I will also talk about our efforts to unravel the conformational differences between two nonallelic mouse insulin and what possibly makes mouse Ins1 a defective isoform. IM-MS coupled with liquid chromatography can detect endogenous pancreatic hormones in samples containing an islet volume equal to one tenth of a single mouse islet. IM-MS provides an orthogonal structural dimension that challenges existing knowledge on essential molecules like insulin and glucagon. Our work aims to accelerate the pace towards development of new therapeutics based on endogenous neuropeptides and hormones.

Thanh Do
University of Tennessee Knoxville
Email: tdo5@utk.edu
volweb.utk.edu/~tdo5
|
 15:40 – 15:45
15:40 – 15:45 |
Interactive Break |
Elucidating Biomolecular Structures and Disease Mechanisms with Ion Mobility Mass Spectrometry
The two predominant challenges in measurement science are to gain structural information of transient but biologically active molecules, and to perform many fast, complete, and accurate sampling of volume-limited samples. I will discuss our recent work using ion-mobility mass spectrometry (IM-MS) to capture a sophisticated model of amyloid dodecamer exhibiting selective cellular toxic vulnerability similar to what have been found in Alzheimer’s disease. I will also talk about our efforts to unravel the conformational differences between two nonallelic mouse insulin and what possibly makes mouse Ins1 a defective isoform. IM-MS coupled with liquid chromatography can detect endogenous pancreatic hormones in samples containing an islet volume equal to one tenth of a single mouse islet. IM-MS provides an orthogonal structural dimension that challenges existing knowledge on essential molecules like insulin and glucagon. Our work aims to accelerate the pace towards development of new therapeutics based on endogenous neuropeptides and hormones.

Thanh Do
University of Tennessee Knoxville
Email: tdo5@utk.edu
volweb.utk.edu/~tdo5
|
 15:45 – 16:30
15:45 – 16:30 |
Plenary Lecture: Ion Mobility Spectrometry: From Instrumental Advances to Understanding the Intersection between Human Health and the Environment Erin Baker, University of North Carolina
|
Ion Mobility Spectrometry: From Instrumental Advances to Understanding the Intersection between Human Health and the Environment
James N. Dodds1, Kaylie I. Kirkwood2, Rebecca Beres1, Jack Ryan1, Anna Boatman1, Jessie Chappel2, Melanie T. Odenkirk2, Karen Butler2, Erin S. Baker1
To date, the use of mass spectrometry (MS)-based analyses has provided extensive knowledge for many different omic studies, especially due to recent advancements in complex sample extractions, front-end chromatography separation, fragmentations strategies, database construction, and bioinformatics tools. However, numerous analytical challenges still remain in MS studies including the analysis of isomeric molecules, suppression of low abundance species in complex samples, disparities in ionization efficiency, and difficulties in data analysis and molecular annotation. Due to these obstacles, both experimental and computational advancements are still greatly needed to enable better omic measurements. In this presentation, I will showcase how ion mobility spectrometry (IMS) has advanced throughout the last thirty years, resulting in IMS separations with higher sensitivity, throughput, and resolving power. These improvements have enhanced the coupling of IMS with various chromatography and MS platforms and provided 3- and 4-dimensional analyses such as LC-IMS-MS or LC-IMS-CID-MS measurements. Currently, these multidimensional analyses are facilitating the separation of highly similar molecules, increasing the number of observed features, and providing improved identification confidence. Furthermore, applications ranging from clinical analyses to environmental assessments are incorporating these simultaneous separations to enable the in-depth omic evaluations necessary for the complex sample types.
[1] Department of Chemistry, University of North Carolina, Chapel Hill, NC
[2] Department of Chemistry, North Carolina State University, Raleigh, NC

Erin Baker
Associate Professor at the University of North Carolina in Chapel Hill, NC
Email: ebaker@ncsu.edu
https://bakerlab.sciences.ncsu.edu/
|
 16:30 – 16:35
16:30 – 16:35 |
Closing Remarks |
|
 –
– |
All times below are listed in Eastern Time (US)
|
|
 09:00 – 12:00
09:00 – 12:00 |
Day 4: The Future of Measurement Science |
|
 09:00 – 10:30
09:00 – 10:30 |
Advances in Measurement Science Lectureship Award Winners |
|
 09:05 – 09:30
09:05 – 09:30 |
Huangxian Ju: "Electrochemiluminescence Biosensing with Nanoparticles as Emitters" Nanjing University
|
Electrochemiluminescence biosensing with nanoparticles as emitters
Electrochemiluminescence or electrogenerated chemiluminescence (ECL) is a light-emitting process, in which the excited state species are generated via exergonic electron transfer on the working electrode, following the radiative transitions to the ground state. It has become a powerful analytical technique due to its electrochemically controllable excited states, low background interference, and high sensitivity. This report starts with its concept and development history, and introduces the innovative ideas and research contents of our group in ECL biosensing with different nanoparticles as the emitters. In 2004, we proposed the first ECL biosensor based on the intrinsic ECL emission of quantum dots (QDs), which brought a new field in ECL biosensing application of QDs and set off a research hotspot of ECL biosensing with inorganic nanoparticles. An electrogenerated precursor was firstly designed for facile synthesis of highly luminescent QDs in aqueous solution, a coreactant for extremely sensitive anodic ECL of QDs was found, and several methods for ECL detection of biomolecules were developed using different QDs. New mechanisms, including intermediate annihilation, the first ECL energy transfer mechanism, surface unpassivation, self-produced coreactant, coreactant consumption, electron and energy transfer, and target-induced structure switching ECL emission for biosensing and signal amplification were also proposed. Besides QDs, a number of inorganic nanoparticles such as g-C3N4 nanosheets, AuNPs-enhanced g-C3N4, cobalt-based MOFs, electroactive MOFs, luminol or Ru(bpy)32+ loaded nanocomposites, dual-stabilizer-capped Au nanoclusters, and copper doped terbium MOFs were also used as the emitters for design of new ECL biosensing mechanisms and development of ECL detection methods. In recently five years, silole-, cyanovinylene- or Ru(bpy)32+ containing polymer dots (Pdots), AIE-active D-A type Pdots, dual RET-enhanced triple-component Pdots and coreactant-embedded Pdots were designed to enhance the ECL emission for sensitive biosensing and bioimaging analysis of biomolecules, cell surface proteins and multiplex microRNAs. These works will lead the development of ECL bioanalysis.

Huangxian Ju
State Key Laboratory of Analytical Chemistry for Life Science, School of Chemistry and Chemical Engineering, Nanjing University
Email: hxju@nju.edu.cn
https://sklac.nju.edu.cn/hxju_en/main.psp
|
 09:30 – 09:55
09:30 – 09:55 |
Tom Rizzo: "Can Vibrational Spectroscopy Finally Find Its Place in the World of Analytical Mass Spectrometry?" Ecole Polytechnique Fédérale de Lausanne (EPFL)
|
Can vibrational spectroscopy finally find Its place in the world of analytical mass spectrometry?
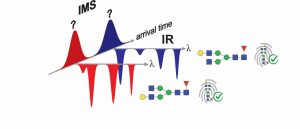
Infrared (IR) spectra of gas-phase ions provide detailed fingerprints that are sensitive to the minutest differences in molecular structure and hence can easily distinguish between isomeric species. Although practiced in many academic research laboratories, IR spectroscopy has not yet found its way into the world of analytical mass spectrometry. There are at least two obvious reasons for this: (1) the addition of a spectroscopic dimension to an analytical measurement has typically taken tens of minutes for each species, making it poorly suited to high-throughput analysis; and (2) the complex, expensive lasers required have made spectroscopic measurements impractical for biomedical research.
We have overcome these problems in an approach that combines ultrahigh-resolution SLIM-based ion mobility, cryogenic IR spectroscopy, and mass spectrometry in a single instrument. By increasing sensitivity and implementing a multiplexing approach to spectral measurement, we can measure an IR fingerprint spectrum of a molecule in as little as 10 seconds. Moreover, we do this using a simple, user-friendly, fiber-pumped IR laser no larger than a shoebox.
After demonstrating the capabilities of our technique, this talk will focus on its application in distinguishing isomeric glycans and glycan-related metabolites. We also have developed schemes to generate IR reference spectra starting from a relatively small number of simple, readily available standards from which we can grow a database for more complex species.

Tom Rizzo
Ecole Polytechnique Fédérale de Lausanne (EPFL)
Email: thomas.rizzo@epfl.ch
https://www.epfl.ch/labs/lcpm/rizzo/
|
 09:55 – 10:20
09:55 – 10:20 |
B. Jill Venton: "Multiplexed Measurements of Neurotransmitters" University of Virginia
|
Multiplexed Measurements of Neurotransmitters
Most electrochemical measurements of neurotransmitters in the brain have been made one at a time at one electrode at a time. In this talk, I will give examples of new strategies to multiplex electrochemical measurements of neurotransmitters, from measuring dopamine and adenosine simultaneously to using two electrodes to measure diffusion. Finally, I will describe new efforts to make both electrochemical and fluorescent based measurements of neurotransmitter simultaneously, allowing different molecules to be detected. Our lab has multiplexed fluorescent G-protein coupled receptors with fast-scan cyclic voltammetry to measure the range of adenosine neuromodulation of dopamine. Multiplexing measurements provides a better picture of complex dynamics of neurotransmitter interactions in the brain.

B. Jill Venton
Professor and Chair of Chemistry Department, University of Virginia
Email: bjv2n@virginia.edu
https://uva.theopenscholar.com/venton-group
Twitter: @jventon
|
 10:30 – 11:15
10:30 – 11:15 |
Session 2: Networking Reception |
|